
H. Nanaocha Sharma
Designation: Scientist-F
Department/Programme Name: Animal Bioresources Division
Phone: 0385-2446122
Academic Qualifications
· 2008-2011, Ph.D, University of Torino, Italy
· 2003-2005, M.Sc. Bioscience, Barkatullah University, Bhopal
Experience
· July, 2024 - till date , Scientist-F, IBSD, Imphal
March, 2019 - July 2024 , Scientist-E, IBSD, Imphal
· April, 2017 - March, 2019, Ramalingaswami Re-entry fellow, IBSD, Imphal
· 2016-2017- Post-Doctoral, Cincinnati Children’s Hospital, Cincinnati, Ohio, USA.
· 2012-2016- Post-Doctoral, Weizmann Institute of Science, Rehovot, Israel
· 2006-2007- First level Bioinformatics, University of Torino, Italy.
Awards & Fellowships
· 2018-2020, Early Career Research Award , DST, Govt. of India
· 2017-2022, Ramalingaswami Re-entry Fellowship, DBT, Govt. of India.
· 2012 - 2014, Marie Curie ITN Fellow (Experience Researcher)
· 2011, Travel Grant for Poster presentation in 5th Mechanobiology Conference at National University of Singapore
· 2006-2007, Italian Government Scholarship
Specialized
Training
· Protein-protein interaction
· Molecular cell biology
1. Research theme photo
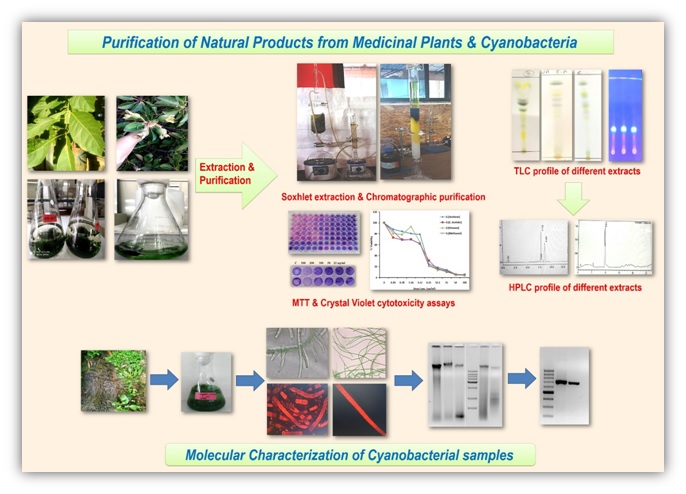
2. Verticals involved
Ethanobotany, Ethnopharmacology & Drug Discovery
3. Research Area/Research Expertise
• Anticancer compounds from diverse bioresources (Medicinal plants & Cyanobacteria) of North East India.
• in vitro Herbal-Drug interaction.
• Cancer Biomarker Discovery and its potential Diagnostic application.
• Genome characterization of Head & Neck cancer from North East India.
4. Research Summary
A. Current Research:
• Potential application of medicinal plants and cyanobacteria from North East India for human health.
Cancer is the leading cause of death globally with approximately 70% from the low and middle income countries. Limitations of the current chemotherapeutic agents is a rising concern which highlights the importance and need for new therapeutic drugs to treat cancer. The vast structural diversity of natural compounds from plants and cyanobacterial species from the biodiversity rich region of the North-East India will serve as a valuable source of new therapeutic drugs. Cyanobacteria are relatively unexplored as potential anticancer agents. Hence assortment, extraction & bioguided fractionation and identification of bioactive compounds from unexplored plants and cyanobacteria for their application in phytopharmaceutical is our main focus of research. Also we aimed to analyse the phytochemical compounds responsible for the anticancer properties and study the molecular mechanism involved using invitro-models.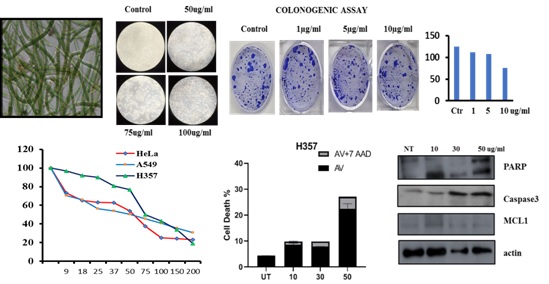
Figure: Cyanobacterial crude extract inhibiting the cell proliferation through apoptosis pathway. Cell cytotoxicity were confirmed by MTT assay and Colonogenic assay. Further cell death by apoptosis were analysed by immunoblotting.
• Quantitative assessment and mapping of the diversity of non-flowering plants (Algae) of Northeast India
Biodiversity assessment is one of the main tools as well as a main goal used in ecology and conservation biology. Microalgae are microscopic, unicellular species that exist solitarily or in chains and are typically found in aquatic systems. They play a vital role in primary production in the aquatic environments and contribute to global atmospheric carbon dioxide acquisition. The commercial value of microalgae is increasing day by day; for example, good nutritional values, large utilization in aquacultures, and a possible use for biofuel production. North East region of India is a genetic treasure house of plants, animals and microbial resources indicating its very rich bioresources. The region forms a distinctive part of the Indo- Burma Biodiversity Hot-Spot which ranks 12th among the 34 Mega Biodiversity Hotspots of the world. Taking the advantage of being at this region, the diversity of non-flowering plants (algae) found in this region will be assessed. In recent years a remarkable growth and interest has been seen in the use of the environmental DNA (eDNA) method as a tool for targeted species detection and biodiversity assessments. Our main focus of research is based on the following objectives:
• Undertake a systematic, quantitative inventory of the distribution and diversity of non-flowering plant (Algae) in the Northeast region using grid-based sampling approach
• Identify algal based biological resources used by local communities in Northeast region and integrate this data into a comprehensive database
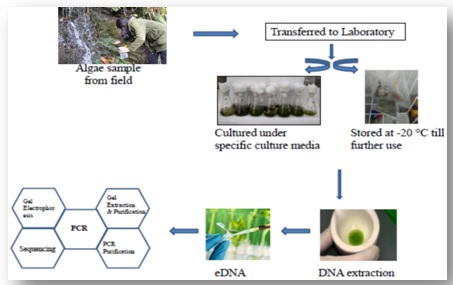
Figure: Schematic representation of methodology followed which involves sampling from
grids, culturing and storage in laboratory for DNA extraction and purity
assessment and molecular characterization for developing a systematic database.
• Development of potential diagnostic and prognostic biomarkers of cancer patients
Manipur, a Northeastern region of India, has highest age-adjusted incidence rates (AARs) of lung cancer (AAR of 26.2/100000 in males and 18/100000 in females) in the country, according to the latest report of National Cancer Registry programme. Lung cancer is a multifactorial disease that results from complex interactions between genetic and environmental factors. Non-small cell lung cancer (NSCLC) is leading cause of cancer related death and the survival rate for patients with NSCLC remain poor so early diagnosis of NSCLC represents the best opportunity for cure. The clinical challenge, therefore, is to develop non-invasive blood biomarkers such as blood-circulating DNA, methylated genes, tumor cells, or micro- RNAs (miRNAs) that may significantly improve the assessment of risk, early diagnosis, and application of chemoprevention for lung cancer.
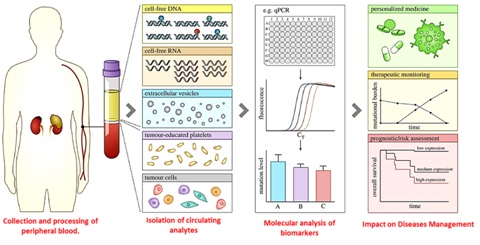
Figure: Circulating tumor cell in Lung cancer: Precision diagnosis and Disease Management.
B. Previous Notable Research:
Role of different domains of p140Cap in tumour supression
p140Cap is an adaptor protein, specifically expressed in brain, testis and epithelial cells that undergoes phosphorylation and tunes its interactions with other regulatory molecules via posttranslational modification to act as tumour suppressor. The major aim of the project was to identify p140Cap regions that could mimic the antitumoral properties of full length p140Cap. We prepared different constructs of p140Cap then transfect them in breast, lung and colon cancer cell line to make stable cell lines. The stable cell lines were used to perform migration assay, in vitro proliferation assay and western blot to study the efficiency of different constructproveds in down regulating the in vitro tumour assay and signaling pathway. Finally, the in vitro results were validated using mice where we did subcutaneous injection of stable cell line expressing different construct of p140Cap. Here, we proved that expression of C-terminal domain of P140Cap can down regulate in vitro and in vivo tumour progression in different tumour cancer cell line (for more information refer Nanaocha sharma et al. 2013).
Investigate how changes in the receptor (outside the cell, or in the transmembrane domain) can invoke differences in signalling occurring in the cytosol of the cell.
Interferon signalling is activated after the formation of ternary complex of interferon and IFNAR1 and IFNAR2 subunits, leading to activation of kinase associated with the cytoplasmic domain. Members of the Type I interferon acts through a single cell surface receptor composing of IFNAR1 and IFNAR2 subunits inducing measurable antiviral and cell growth inhibition. Interestingly, very different signaling programs can be initiated by interferons, depending on the interferon subtype, its concentration and time of activation. To investigate how this is possible, we combine biophysical work with cell biology. Here we investigated how changes on the outside of the cell are transmitted inside. By inserting 1-5 alanine residues at either the C or N-terminal region of the transmembrane domain of IFNAR1, we are causing two movements of the extracellular domain in relation to the interacellular domain. Each alanine is rotating the helix by 1000 and translating it 1.5Å along the helical axis. After transfection to cells lacking endogenous IFNAR1 we measured IFN binding, basal and IFN induced STAT phosphorylation and robust and tunable gene induction. The main conclusion from the results obtained is that insertion of alanine within the TMD does not produce any detectable change in downstream signaling. Therefore, we can assume that rotating the alpha helix within the TMD of IFNAR1, or its translation has no effect on the biological activity of the receptor compared to wild type. While these experiments clearly show plasticity between the extracellular and intracellular domains, they do not provide a direct indication which of them is affected by the induced insertions. To this end we mutated residues at the transmembrane–cytosolic junction modulates dimerization and affects activation. However, for IFNAR1 such mutations again fail to demonstrate any effect (either quantitative of qualitative) in biological readouts. On the extracellular side of the TMD, we then added 2 to 10 serine amino-acids, affecting the location of the extracellular domain of IFNAR1 in relation to IFNAR2. Only 10 serine residues weakened IFN binding to the surface by ~10 fold. As we do not observe IFN binding in the absence of IFNAR1, we can assume that IFN promotes the complexation of the S10 IFNAR1 mutant with IFNAR2, however to a lesser extent than the wild-type. Finally, we wanted to see whether the sequence of the TMD of IFNAR1 is at all important for signaling. Therefore, we replaced the TMD of IFNAR1 with that of IFNAR2, practically having the same TMD sequence on both receptors. Again, this had no effects on either IFN binding or signaling. These results differ from the findings for cytokines where downstream signaling was related to the homo-dimerization of the TMDs. Our findings are surprising, as they clearly demonstrate that the relative orientation and translation of both the intra and extracellular domains of IFNAR1 and IFNAR2 are of no importance for signaling. Basically, this comprehensive study suggests that role of interferon is to bring IFNAR1 and IFNAR2 into proximity for a given time, and that this proximity is sufficient to drive signaling. This simplistic view of events is in line with our previous finding that binding affinity of interferons is the main determinant that dictates differential activities of type I IFNs (for more information please refer Nanaocha Sharma et al. 2015, JBC).
In another project we investigate the relation between receptor plasticity and signaling. Domain movements of interferon receptor have multiple effects on signaling. One effect would be to transform the apo towards the holo state of the receptor, with holo state being of higher binding affinity and thus promoting signaling. A second effect would be to transform a non signaling complex to an active one. In this case, dimerization of the two interferon receptor subunits is not sufficient to induce signaling, which depends on structure plasticity and domain movements. It has been proved that substituting SD4 domain of IFNAR1 with a corresponding domain from homologous cytokine receptors did not restore high affinity ligand binding, suggesting that signaling involves specific communication from the extracellular domain inwards. Here, we propose to investigate the relationship between receptor flexibility, domain movement and signaling upon interferon binding. In order to restrict the receptor flexibility and domain movement, we introduced single and multiple disulfide bridge between different domains of the IFNAR1. Biological responses upon interferon binding were determined in a HUH7 cell line lacking the endogenous IFNAR1. In addition, the extracellular domains of the mutant receptors were expressed in insect cells and IFN binding was determined using SPR. Introduction of disulfide bonds between SD3 and SD4 domains resulted in a significant increase in antiviral propensity, stronger induction of STATs and higher gene upregulation than the wild-type clones. These results were in accordance with our in vitro binding measurements. Conversely, linking together the other domains decreases binding and biological activity.
Clathrin-dependent endocytosis of the type I interferon (IFN-?/?) receptor (IFNAR).
Here, we showed that the internalized IFNAR1 and IFNAR2 subunits of the IFNAR complex are differentially sorted by the retromer at the early endosome. Binding of the retromer VPS35 subunit to IFNAR2 results in IFNAR2 recycling to the plasma membrane whereas IFNAR1 is sorted to the lysosome for degradation. Depletion of VPS35 leads to abnormally prolonged residency and association of the IFNAR subunits at the early endosome resulting in increased activation of STAT1 and IFN-dependent genes transcription. These experimental data establish the retromer complex as a key spatiotemporal regulator of IFNAR endosomal sorting and a new player in IFN-induced JAK/STAT signaling and gene transcription.
Research Articles (Peer Reviewed, Only authorized publishers) – Recent First
1. Mohanta YK, Panda, SK, Jayabalan R, Sharma N, Bastia AK, Mohanta TK (2017). "Antimicrobial, Antioxidant and Cytotoxic Activity of Silver Nanoparticles Synthesized by Leaf Extract of Erythrinasuberosa (Roxb.)." Front Mol Biosci 4: 14. PubMed ID: 28367437
2. Li, H., Sharma, N., General, I. J., Schreiber, G., & Bahar, I. (2017). Dynamic Modulation of Binding Affinity as a Mechanism for Regulating Interferon Signaling. Journal of molecular biology, 429(16), 2571–2589. PubMed ID:28648616
3. Grasso S, Chapelle J, Salemme V, Aramu S, Russo I, Vitale N, Verdun di Cantogno L, Dallaglio K, Castellano I, Amici A, Centonze G, Sharma Nanaocha, Lunardi S, Cabodi S, Cavallo F, Lamolinara A, Stramucci L, Moiso E, Provero P, Albini A, Sapino A, Staaf J, Di Fiore PP, Bertalot G, Pece S, Tosoni D, Confalonieri S, Iezzi M, Di Stefano P, Turco E, Defilippi P. (2017). The scaffold protein p140Cap limits ERBB2-mediated breast cancer progression interfering with RacGTPase-controlled circuitries. Nat Commun. 16;8:14797. PubMed ID: 29600801
4. Chmiest D, Sharma Nanaocha, Zanin N, Viaris de Lesegno C, Shafaq-Zadah M, Sibut V, Dingli F, Hupé P, Wilmes S, Piehler J, Loew D, Johannes L, Schreiber G, Lamaze C. (2016). Spatiotemporal control of interferon-induced JAK/STAT signalling and gene transcription by the retromer complex. Nat Commun. 5;7:13476. PubMed: 27917878
5. Sharma Nanaocha, Longjam G, Schreiber G. (2016). Type I Interferon Signaling is decoupled from Specific Receptor Orientation through Lenient Requirements of the Transmembrane Domain. J Biol Chem. 12;291(7):3371-84. PubMed ID: 26679999
6. Urin V, Levin D, Sharma Nanaocha, Harari D, Schreiber G. (2015). Fine Tuning of a Type 1 Interferon Antagonist. PLoS One. Jul 9;10(7):e0130797. PubMed ID:26158644
7. Levin D, Schneider W.M, Hoffmann H-H, Yarden G, Busetto A.G, Manor O, Sharma Nanaocha, Rice C.M, Schreiber G. (2014). Multifaceted Activities of Type I Interferon Are Revealed by a Receptor Antagonist. Science Signaling. 07(327), ra50. PubMed ID: 24866020
8. Sharma Nanaocha, et al “Identification of two regions in the p140Cap adaptor protein that retain the ability to suppress tumor cell properties”. (2013). Am. J Cancer Res. 3(3):290-301. PubMed ID: 23841028
9. Di Stefano P, Camacho Leal M.P, Tornillo G, Bisaro B, Repetto D, Pincini A, Santopietro E, Sharma Nanaocha, Turco E, Cabodi S, Defilippi P. (2011). The adaptor proteins p140CAP and p130CAS as molecular hubs in cell migration and invasion of cancer cells. Am. J Cancer Res. 1(5):663-673. PubMed ID: 21994904
10. Di Stefano P, Camacho Leal M. P., TornilloG,Bisaro B, Damiano L, Morello V, RepettoD,Pincini A, Aramu S, Sharma Nanaocha, Grasso S, Turco E, Cabodi S, Defilippi P. (2011). p140Cap and p130Cas Adaptor Proteins as Molecular Hubs in Cell Migration and Invasion. Connective Tissue Research. 52(4): 255–289. PubMed ID: 21994904
11. Repetto D, Aramu S, BoeriErba E, Sharma N, Grasso S, Russo I, Jensen ON, Cabodi S, Turco E, Di Stefano P, Defilippi P. (2013). Mapping of p140Cap phosphorylation sites: the EPLYA and EGLYA motifs have a key role in tyrosine phosphorylation and Csk binding, and are substrates of the Abl kinase. PLoS one. 8(1):e54931.
PubMed ID: 23383002

Group Members (Details):
|
1. |
Heisnam Rameshwari Devi PhD JRF, ICMR Email ID: rameheisnam@gmail.com Extraction and molecular study of natural compounds from fresh water cyanobacteria Tolypothrix sp. for their anticancer activity. Expected Outcome: Anticancer activity of cyanobacteria of effective cell death and pathway analysis. |
|
|
2. |
Khaidem Devika Chanu PhD JRF, CSIR Email ID: demoky11may@gmail.com
Phytochemical analysis of NE (North-eastern) medicinal plants for its anti-tumor properties against different cancer cell lines.
Expected Outcome: Deciphering the molecular mechanisms involved in cell apoptosis or cytotoxicity. |
|
|
3. |
Soibam Thoithoisana Devi PhD JRF, CSIR Email ID: thoi2sana@gmail.com Screening of Medicinal plants and Fresh Water Cyanobacteria as Anticancer Drug Resource.
Expected Outcome: Finding Bioactive compounds which have anti proliferative activity against different cancer cell lines. |
|
|
4. |
Kshetrimayum Vimi PhD JRF, IBSD Email ID: : huk7vimi@gmail.com Uncovering Anticancer agents from diverse Natural Origin and determination of the apoptosis pathway. Expected Outcome: Identification of bioactive compounds and elucidation of the cytotoxic pathways. |
|
|
5. |
Hawaibam Monalisha Project JRF, DBT Email ID: monalisahawaibam@yahoo.in Project Title : “Design, Synthesis and Modelling of Type I Interferon Antagonist for treatment of Tuberculosis” Isolation and purification of interferon proteins, induction of random mutation and study of receptor protein binding affinity. Expected outcome: Isolation of interferon proteins, sequencing of the plasmid construct and carry out large scale synthesis of interferon protein for further study. |
|
|
6. |
Heisnam Sobhana Devi Project Fellow, DBT Email ID: livyaheisnam@gmail.com Project Title: Quantitative assessment and mapping of the diversity of non-flowering plants (Algae) of North East India Inventory of the distribution and diversity of Algae in Northeast India. Expected outcome: Repository and genetic database of algal diversity and distribution in Northeast India. |
|
|
7. |
Nepuni Rinaldi Project Fellow, DBT Email ID: rinaldinepuni03@gmail.com Project Title: Quantitative assessment and mapping of the diversity of non-flowering plants (Algae) of North East India Inventory of the distribution and diversity of Algae in Northeast India. Expected outcome: Repository and genetic database of algal diversity and distribution in Northeast India. |
|
|
8. |
Kamaljit Moirangthem, PhD Project RA, DBT Email ID: kamaljit.moirangthem@hotmail.com Project Title: Quantitative assessment and mapping of the diversity of non-flowering plants (Algae) of North East India Inventory of the distribution and diversity of Algae in Northeast India. Expected Outcome: Repository and genetic database of algal diversity and distribution in Northeast India. |
|
|
9. |
Deepak Singh Lourembam, PhD DBT-RA (Post-Doctoral) Email ID: deelourembam@gmail.com Title: Development of potential diagnostic tools of Lung cancer patients using blood miRNAs and DNA methylation of cancer specific genes and circulating cell free DNA Expected Outcome: Screening of Circulating cell free DNA content in Healthy and Lung Cancer patients for determination of potential diagnostic value. |
|
|
10 |
Keithellakpam Ojit Singh, PhD Scientist Fellow, IBSD Email ID: ojit1999@gmail.com Research Interest: Value-added products, Bioactive compounds Expected outcome: Screening of cyanobacteria at Algal Repository for bioactive compounds for anti-cancer activities. |
|
1. Extramural:
a. Present:
|
Sl. No. |
Duration (From-To) |
Title of Project/ Grant |
PI/Co-PI |
Funding Agency |
|
|
1. |
2020-2023 |
Genome India: Cataloguing the genetic variation in Indians. |
PI |
DBT |
|
|
2. |
2018-2020 |
Quantitative assessment and mapping of the diversity of non-flowering plants (Algae) of North East India |
PI |
DBT |
|
|
3. |
2018-2020 |
Design, synthesis and modelling of Type 1 interferon antagonist for treatment of Tuberculosis. |
PI |
DST |
|
|
4. |
2017-2022 |
Engineering type I interferon that binds with different affinity to the receptor which modulate biological read out. |
PI |
DBT |
|
POSITIONS
Students with M.Sc. or PhD degree in Life science are welcome to apply to our Ph.D. and post doctoral are encourage to apply in different projects going on in our lab. Candidates with experience in molecular biology, cell biology, biochemistry and bioinformatics will be preferred.
Interested candidates should contact me at sharma[dot]nanaoch[at]gmail.com










