
Sudripta Das
Plants
Designation/Position: Scientist-F
Department/Programme Name: Plant Bioresources
Phone: 0385-2446122, Ext: 213
Academic Qualifications
• 2011-12: Post-Doctoral Fellow in Noble Research Centre, Oklahoma State University, under Prof. Ramanjulu Sunkar
• 2001-2004: IFCPAR-CEFIPRA Post-Doctoral Fellowship and worked under Prof. David Tepfer at Lab. Rhizosphere, INRA, France
• 1992-1999: PhD, University of Calcutta, Kolkata under Prof. Sumita Jha
• 1990-1991: MSc Botany, University of Calcutta, Kolkata
Experience
• Presently: Scientist-F, IBSD, Imphal
• June 2013: Scientist-E, IBSD, Imphal
• February 2013-June 2013: Scientist-E, Tocklai Tea Research Institute, Jorhat
• April 2008-January 2013: Scientist-D, Tocklai Tea Research Institute, Jorhat
• January 2004-March2008: Scientist-C, Tocklai Tea Research Institute, Jorhat
Awards & Fellowships
• IFCPAR-CEFIPRA Post-Doctoral Fellowship in 2001
• DBT-CREST Award 2011
Memberships (Professional Associations / Societies)
• Member of International Society for Plant Molecular Biology (ISPMB)
• Member of International Association for Plant Tissue Culture & Biotechnology (IAPTC & B)
PhD’s Guided
Name Title of thesis University Status
Bornali Gohain Designing a quality index for elite Guwahati Univ Awarded
Camellia sinensis clones based on 2012
molecular and gene expression studies
Sushmita Gupta Study of Transcriptome Polymorphisms Guwahati Univ Awarded
and development of molecular markers 2012
associated with drought tolerance for
marker assisted selection in Camellia sinensis
Sourabh K Das Analysis of a Polyploid Population in Guwahati Univ Awarded
Tea Using Structural/ Functional 2013
Genomic Techniques
Priyadarshini Bhorali Identification of Molecular Markers Guwahati Univ Awarded
Associated with Blister Blight 2013
(Exobasidium vexans) Disease in
Camellia sinensis
Tirthankar Identification of differentially Guwahati Univ Awarded
Bandyopadhyay expressed genes induced by infestation 2012
with Helopeltis theivora in Camellia sinensis
(L.) O. Kuntze
Raju Bharalee Generation, analysis and identification Guwahati Univ Awarded
of Expressed Sequence Tags of differentially 2012
expressed genes of Camellia sinensis
(L.) O. Kuntze under abiotic stress
(drought) using suppression subtractive hybridization
Niraj Agarwala Study and analysis of differentially Guwahati Univ Awarded
expressed genes in tea (Camellia 2015
sinensis (L).O.Kuntze) in response to
Exobasidium vexans infection
H. Ranjit Singh Transfer and analysis of defense Guwahati Univ Awarded
genes for conferring resistance against 2015
blister blight in Camellia sinensis (L).O.Kuntze
Parveen Ahmed Development of Molecular Markers Guwahati Univ Awarded
for Helopeltis resistance in Tea 2014
Prasenjit Bhagwati Study of genetic variability related to Guwahati Univ Awarded
pigmentation and flavour formation in 2015
wild and cultivated tea germplasms
Neelakshi Decaffeinated Tea: Through Guwahati Univ Awarded
Bhattacharyya Gene Silencing 2016
Other Achievements
• Member of Scientific Advisory Committee (SAC) of National Institute of Malarial Research (NIMR-ICMR)
1. Research theme photo
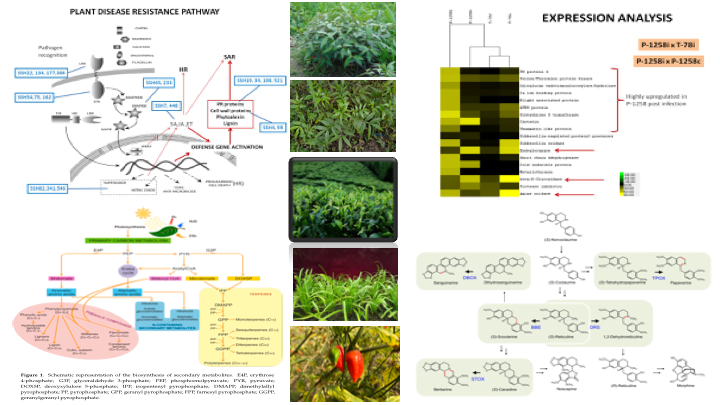
2. Verticals involved
Plant Resources
3. Research Area/Research Expertise
• Plant Stress Biology
• Molecular mechanisms of plant secondary metabolite biosynthetic pathways
4. Research Summary
A. Current Research:
TITLE: Functional Genomic Analysis of Secondary Metabolic Pathways for Production of Berberine alkaloids and Flavonoids
In the post-genomic era, integrated analysis of comprehensive gene expression (transcriptome) and metabolic profiling (metabolome) has been successful in plant functional genomics. Deep transcriptome technology, also known as RNA-seq, provides whole transcriptome expression profiles of selected plant tissues or cells, thereby permitting the integrated analysis of transcriptomics and metabolomics in any plant species. The present study would utilize deep transcriptomics combined with targeted metabolic profiling to identify candidate genes and factors associated with secondary metabolite production of berberine alkaloids and catechins. The obtained spatial and temporal datasets would be useful resources for further studies involving intermediate metabolites and novel compounds.
The application of systematic genome mining approaches in combination with functional analysis of candidate clusters offers enormous potential for unravelling of pathways, enzymes and chemistries related to production of berberine alkaloids and flavonoids from plants like Berberis sp./Zanthoxylum sp. and Polygonum sp. respectively.
B. Previous Notable Research:
TITLE: Allele Mining for Stress Resistance in Tea (DBT-Unit of Excellence)
To understand the gene networks that underlie tea stress and defense responses, it is necessary to identify and characterize the genes that respond both initially and as the physiological response to the stress or pathogen develops. We used PCR-based suppression subtractive hybridization to identify Camellia sinensis genes that are differentially expressed in response to drought, fungal pathogen and infestation by green flies and thrips. We identified a total of 19,231 differentially expressed genes from sixteen stress cDNA libraries. RT-PCR analysis revealed that 45% of the stress-inducible genes are rarely transcribed in unstressed plants and 10% of them were not previously represented in Camellia expressed sequence tag databases. More than two-thirds of the genes in the stress cDNA collection have not been identified in previous studies as stress / defense response genes. Several stress-responsive cis-elements showed a statistically significant over-representation in the promoters of the genes in the stress cDNA collection. These include W-, F- and G-boxes, the JA inducible element, the abscisic acid response element and cysteine rich motif. The stress cDNA collection comprises a broad repertoire of stress-responsive genes encoding proteins that are involved in both the initial and subsequent stages of the physiological response to abiotic stress and pathogens. The degradome analysis using RNA-seq data suggests that regulation of the transcriptome is universally governed by specific small RNA target molecules which may be modulated. This set of stress-, pathogen- and positively regulated tea quality related genes is an important resource for understanding the genetic interactions underlying stress signaling and responses and may contribute to the characterization of the stress transcriptome through the construction of standardized specialized arrays.
> For drought tolerance, Koomsung 29 was identified as the most tolerant among the germplasms so far characterized, which is also well characterised for its quality.
> For blister blight tolerance, Phoobshering 1278 was resistant to blister blight disease and should be utilised for breeding of Darjeeling clones.
A. Research Articles – Relevant publications
1. “Transgenic Tea Over-expressing Solanum tuberosum Endo-1,3-beta-D-glucanase Gene Conferred Resistance Against Blister Blight Disease” H. Ranjit Singh, Pranita Hazarika, Manab Deka & Sudripta Das. Plant Molecular Biology Reporter (2018) 36:107–122 https://doi.org/10.1007/s11105-017-1063-x.
2. “A Proteomic Approach to Evaluate the Effects of Endogenous Expression of Cryptogein Gene in Crypt Transgenic plants of Bacopa monnieri” J. Appl. Biotechnol Bioeng. (2017) 4(3): 100-104.
3. “Construction of a genetic linkage map and mapping of drought tolerance trait in Indian beveragial tea” Sapinder Bali, Vishnu Bhat, A Mamgain, Sudripta Das, S N Raina, A K Pradhan, S K Yadava and S Goel. Mol Breeding. 2015. 35:112. [DOI 10.1007/s11032-015-0306-5].
4. “Enhanced resistance to blister blight in transgenic tea (Camellia sinensis [L.] O. Kuntze) by overexpression of class I chitinase gene from potato (Solanum tuberosum)”. H. Ranjit Singh, Manab Deka and Sudripta Das. Funct Integr Genomics. 2015. [DOI 10.1007/s10142-015-0436-1].
5. “Molecular Landscape of Helopeltis theivora Induced Transcriptome and Defense Gene Expression in Tea”. Tirthankar Bandyopadhyay, Bornali Gohain, Raju Bharalee, Sushmita Gupta, Priyadarshini Bhorali, Sourabh Kumar Das, M C. Kalita and Sudripta Das. Plant Mol Biol Rep. 2014. [DOI 10.1007/s11105-014-0811-4].
6. “Molecular Analysis of Drought Tolerance in Tea by cDNA-AFLP Based Transcript Profiling”. (2013) Sushmita Gupta, Raju Bharalee, Priyadarshini Bhorali, Sourabh K. Das, Prasenjit Bhagawati, Tirthankar Bandyopadhyay, Bornali Gohain, Niraj Agarwal, Parveen Ahmed, Sangeeta Borchetia, M. C. Kalita, A. K. Handique, Sudripta Das. Mol. Biotechnol. 53:237–248 [DOI: 10.1007/s12033-012-9517-8].
7.“Biochemical evaluation…….. catechin rich genotypes” (2013). S. K. Das, S. Sabhapandit, G. Ahmed and Sudripta Das. Biochem Genetics. June 2013, Volume 51, Issue 5-6, pp 358-376 [DOI: 10.1007/s10528-013-9569].
8. “Identification of Differentially……………Suppression Subtractive Hybridization” A Das, Sudripta Das and T K Mondal. (2012). Plant Mol Biol Rep. 30:1088–1101.
9. “Understanding Darjeeling Tea Flavour on a Molecular Basis” Bornali Gohain, Sangeeta Borchetia, L.P. Bhuyan, A. Rahman, K. Sakata, M. Mizutani, B. Shimizu, G. Gurusubramaniam, R. Ravindranath, M. Hazarika and Sudripta Das. Plant Mol Biol. 78: 577-597 (doi:10.1007/s11103-012-9887-0). 2012.
10. “Identification of drought tolerant progenies in tea by gene expression analysis”. Sushmita Gupta, Raju Bharalee, Priyadarshini Bhorali, Tirthankar Bandyopadhyay, Bornali Gohain, Niraj Agarwal, Parveen Ahmed, Hemanta Saikia, Sangeeta Borchetia, MC Kalita, AK Handique and Sudripta Das. Funct Integr.Genomics (2012). 12: 543-563. [DOI: 10.1007 /s10142-012-0277-0].
B. Books Authored – Recent First
Sushmita Gupta and Sudripta Das. Gene Networks Involved In Water Deficit Stress Response In Tea. 2012. Pages 264. LAP LAMBERT Academic Publishing. ISBN: 978-3-659-29025-1.
C. Book Chapters – Recent First
1. Sudripta Das, S.C. Das and M. Hazarika. Global Tea Breeding: Achievements, Challenges and Perspectives - Breeding of the Tea Plant (Camellia sinensis) in India. (2012). Springer-Zhejiang University Press.
2. Sudripta Das and Sumita Jha. Tissue culture of Cashew Nut. 2003. In: Prof. S. S. Bhojwani Commemorative Volume. Springer-Verlag.
3. Sudripta Das and Sumita Jha. Strategies for improvement of Cashewnut through tissue culture. 1-7 (1995). In: Plant Tissue Culture, Islam, A.S. (ed), Oxford-IBH.
|
1. |
Dr. Roseeta Mutum, DBT-PDF Email: roseeta.5@gmail.com Problem Addressing: Investigating and integrating the role of microRNAs in metabolite pathway of ethno-medicinally important plant Allium hookeri |
|
|
2. |
Ms. Pukhrambam Premi Devi, Senior Research Fellow Email: premi.pukhrambam@gmail.com Problem Addressing: Gene regulation study of flavonoid biosynthetic pathway in Polygonum posumbu |
|
|
3. |
Ms. Moirangthem Lakshmipriyari Devi, Junior Research Fellow Email: lakshmipriyaridevi@gmail.com Problem Addressing: Genomic and Biochemical Analysis of berberine and sanguinarine production in Zanthoxylum species of Manipur |
|
|
4. |
Ms. Kh. Khedashwori Devi, Junior Research Fellow Email: khkhedashwori@gmail.com Problem Addressing – Characterization of Capsicum sp. of NE India for value addition |
|
|
5. |
Ms. Khundrakpam Basanti, Laboratory Assistant Email: buntylive@yahoo.com |
|
A. Core/Internal
a. Present:
|
Sl. No. |
Duration (From-To) |
Title of Project/ Grant |
PI/Co-PI |
Funding Agency |
|
|
1 |
2014-present |
Functional Genomic Analysis of secondary metabolite pathways for production of flavonoids and berberine alkaloids |
Sudripta Das (PI) |
IBSD |
|
b. Past:
|
Sl. No. |
Duration (From-To) |
Title of Project/ Grant |
PI/Co-PI |
Funding Agency |
|
|
1 |
2017-2019 |
Demonstration and Capacity Building Programmes for Production of Naga Chilies by Modern Horticulture Technology at Farmers Field in Nagaland |
Sudripta Das (PI) |
IBSD |
|
|
2 |
2017-2019 |
Demonstration of Sustainable Technologies for Protected and Open Cultivation of Important Vegetable Crops in Farmers Field and IBSD Mizoram |
Sudripta Das (PI) |
IBSD |
|
B. Extramural:
a. Present:
|
Sl. No. |
Duration (From-To) |
Title of Project/ Grant |
PI/Co-PI |
Funding Agency |
|
|
1. |
2019-2024 |
Conservation, propagation and mass multiplication of selected orchid species for developing bio-based entrepreneurship in North-East India |
Co-Principal Investigator |
DBT, GoI |
|
|
2. |
2020-2015 |
Earth BioGenome |
Principal Investigator |
DBT-International Funding |
|
b. Past:
|
Sl. No. |
Duration (From-To) |
Title of Project/ Grant |
PI/Co-PI |
Funding Agency |
|
|
1 |
2013-2017 |
Allele Mining for Stress Resistance in tea |
Principal Investigator |
DBT-Unit of Excellence |
|
|
2 |
2011-2014 |
Identification and characterization of sequence based markers for genetic improvement of tea |
Principal Investigator |
DBT (GoI) |
|
|
3 |
2010-2013 |
Molecular documentation of Darjeeling Tea clones using DNA profiling |
Principal Investigator |
DBT (GoI) |
|
|
4 |
2010-2014 |
Tea Bioinformatics |
Principal Investigator |
MICT-DICT |
|
|
5 |
2010-2013 |
Establishment of Institutional Biotech Hubs (IBT Hubs) by DBT under special programme for North Eastern States of India |
Principal Investigator |
DBT-NERBPMC |
|
|
6 |
2008-2013 |
Abiotic and Biotic Stress Analysis for development of Stable Quality Genotypes |
Principal Investigator |
Tea Board 11th Plan |
|
|
7 |
2006-2009 |
Characterization and enhancement of volatile flavour components and gene expression profiling in Darjeeling black tea infested by green fly and thrips |
Principal Investigator |
INDO-JAPAN Collaborative Project |
|
|
8 |
2004-2009 |
Developing production system for tea polyphenols and their condensed products |
Principal Investigator |
CSIR-NMITLI |
|
|
9 |
2007-2010 |
Production of terpenoids in untransformed and transformed cell and organ cultures in tea |
Principal Investigator |
DBT (GoI) |
|
|
10 |
2007-2010 |
Decaffeinated Tea: Through Gene Silencing |
Principal Investigator |
DST (GoI) |
|
|
11 |
2007-2010 |
Development of transgenics for blister blight resistance in tea |
Co-Principal Investigator |
DBT (GoI) |
|
|
12 |
2006-2010 |
Development of integrated genetic linkage map and marker assisted selection in tea |
Principal Investigator |
DBT (GoI) |
|
|
13 |
2006-2010 |
Generation and analysis of Expressed Sequence Tag (EST) from Camellia species |
Principal Investigator |
DBT (GoI) |
|





